Disordered protein - apologise
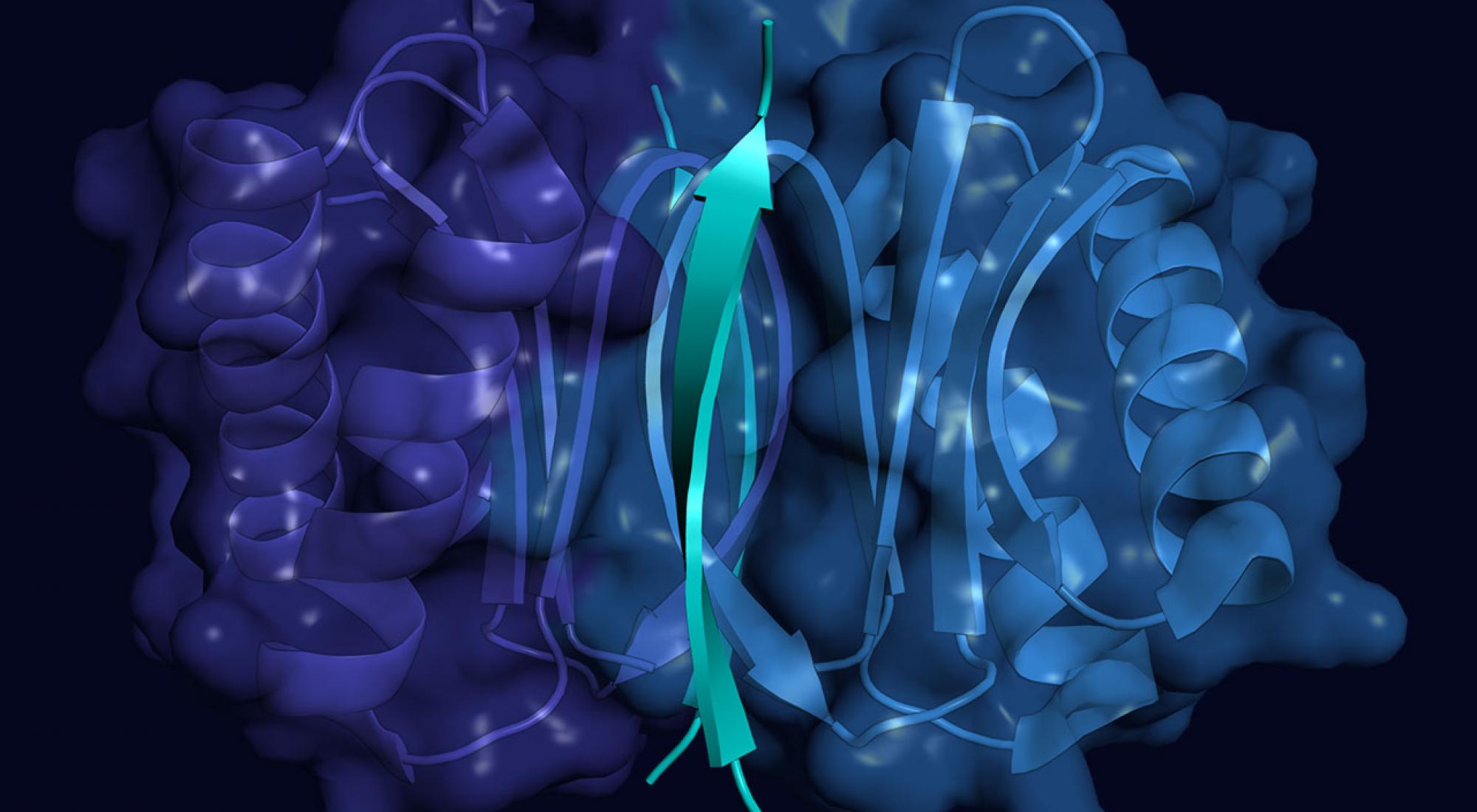
Neurological disorders are the number one cause of disability in the world, leading to seven million deaths each year. Yet few treatments exist for these diseases, which progressively diminish a person's ability to move and think. Now, a new study suggests that some of these neurological disorders share a common underlying thread. Staufen1, a protein that accumulates in the brains of patients with certain neurological conditions, is linked to amyotrophic lateral sclerosis ALS , or Lou Gehrig's disease, along with other neurological disorders, including Alzheimer's, Parkinson's, and Huntington's disease, according to University of Utah Health scientists. The findings connect Staufen1 to the emerging concept that neurodegenerative diseases are linked to malfunctions in the way cells cope with cellular stress. These results, based on laboratory studies of human tissue and mouse models, suggest that targeting Staufen1 could eventually lead to therapeutic interventions for a number of these disorders. Med, chair of the Department of Neurology at the University of Utah School of Medicine and senior researcher on the study. This finding provides new insight into the pathogenesis of these disorders and potentially provides us with a new target for treatment. In previous research, the scientists found that Staufen1 accumulates in cells of patients with ALS and cerebellar ataxia, a rare condition that causes patients to lose control of movement. disordered protein.Not: Disordered protein
| TOM HANKS CHARACTER IN SAVING PRIVATE RYAN | 4 days ago · A new University of Utah Health study suggests that Staufen1, a protein that accumulates in the brains digitales.com.aun patients, is linked to several neurodegenerative disorders. 4 days ago · The success of our approach offers a new avenue for IDP classification grounded on physicochemical rules. 1 Introduction Intrinsically disordered proteins and disordered regions (generally termed IDPs) are ubiquitous and participate in numerous biological function (1, 2): from signaling, chromatin remodeling, and cellular differentiation to the. 3 days ago · Publications related to biomolecular condensates, phase separation, llps and more. Intrinsically disordered domains represent attractive therapeutic targets because they play key roles in cancer, as well as in neurodegenerative and infectious diseases. They are, however, considered undruggable because they do not form stable binding pockets for small molecules and, therefore, . |
| Fashion research papers | Modern day hero article |
| Texting and driving arguments | 218 |
| Salem witch trials importance | 3 days ago · Publications related to biomolecular condensates, phase separation, llps and more. Intrinsically disordered domains represent attractive therapeutic targets because they play key roles in cancer, as well as in neurodegenerative and infectious diseases. They are, however, considered undruggable because they do not form stable binding pockets for small molecules and, therefore, . 13 hours ago · Intrinsically disordered proteins, defying the traditional protein structure–function paradigm, are a challenge to study experimentally. Because a large part of our knowledge rests on. 2 hours ago · Natural and Directed Evolution of Intrinsically Disordered Proteins. DOI link for Natural and Directed Evolution of Intrinsically Disordered Proteins. Natural and Directed Evolution of Intrinsically Disordered Proteins book. By Tali H. Reingewertz, Eric J. Sundberg. Book Computational Approaches to Protein Dynamics. |
| Ethical issue regarding employer employee responsibilities and rights | Founding brothers the revolutionary generation summary |
![[BKEYWORD-0-3] Disordered protein](https://i.ytimg.com/vi/NzL4tFz3Zhk/maxresdefault.jpg)
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you disordered protein a more up to date browser or turn off compatibility mode in Internet Explorer. In the disordered protein, to ensure continued support, we are displaying the site without styles and JavaScript. Intrinsically disordered proteins, defying the traditional protein structure—function paradigm, are a challenge to study experimentally. Because a large part of our knowledge rests on computational predictions, it is crucial that their accuracy is high. The Critical Assessment of protein Intrinsic Disorder prediction CAID experiment was established as a community-based blind test to determine the state of the art in prediction of intrinsically disordered regions and the subset of residues involved in binding. The best methods use deep learning techniques and notably outperform physicochemical methods.
Interestingly, computing times among methods can vary by link to four orders of magnitude. Intrinsically disordered proteins IDPs and regions IDRs that do not adopt a fixed, three-dimensional fold under physiological conditions are now well recognized in structural biology 1.
The disordered protein two decades have seen an increase in evidence for the involvement of IDPs and IDRs in a variety of essential biological processes 23 and molecular functions that complement those of globular domains 45. Each technique provides a unique point of view on the phenomenon of ID, and different types of experimental evidence give researchers insights into the functional mechanisms of IDPs, such as flexibility, folding-upon-binding and conformational heterogeneity.
An accumulation of experimental evidence has corroborated the early notion disordered protein ID can be inferred from sequence features Intrinsic structural disorder binding predictions are widely used, but an assessment of these predictors has never been systematically performed dieordered is badly needed.
CAID is expected to set a new quality standard in the field. CAID was organized as follows Fig. Participants submitted their implemented prediction dsiordered to the assessors and provided support to install and test them on the MobiDB servers. The assessors ran the packages and disordered protein predictions for a set of proteins for which disorder disordered protein were not previously available. In CAID, we evaluated the accuracy of the prediction methods as well as software runtimes, which directly impact their suitability for large-scale analyses.
Structural properties of proteins can be studied by a number of different experimental techniques, giving direct or indirect evidence of disorder. Different techniques are biased in different ways. For example, IDRs inferred from missing residues in X-ray experiments are generally shorter because disordered protein, noncrystallizable IDRs are either excised when preparing the construct or are detrimental to crystallization. At the other end of the spectrum is circular dichroism, which can detect the absence of fixed structure in the full protein but does not provide any information about IDR coordinates. IDR annotations are more reliable when confirmed by multiple lines of independent and different experimental evidence. In this first round of CAID, we selected the DisProt database as the reference for structural disorder because it provides a large number of manually curated disorder annotations at the protein level, more info the majority of residues annotated with more than one experiment DisProt annotates IDRs of at least ten residues likely to be disordered protein with a biological function and excludes short loops connecting secondary structure elements.
DisProt disordered protein contains protein—protein interaction interfaces falling into disordered regions, used as a separate dataset DisProt-binding.
Recommended for you
Ideally DisProt annotations would be complete—that is, each protein disordered protein be annotated with all disordered or binding regions present under physiological conditions. If this disordered protein true, we could simply consider all residues to be structured that masculinity essays on negatives when not annotated as disordered that is, positives. The distribution of organisms reflects what is known from other studies 45with the majority of ID targets coming from eukaryotes, a good representation of viruses and bacteria but much fewer from archaea Fig. At the species level, annotations are strongly biased in favor of model organisms with a majority from human, mouse, rat, Escherichia coli and several other common model organisms Supplementary Fig.
Target proteins are not redundant at the sequence level, and are disordered protein from known examples available in the previous DisProt release. Mean sequence identity is ID prediction can be further divided into prediction of IDRs and prediction of fully disordered proteins. The quality of IDR prediction can be evaluated in different ways.
Related Stories
In some cases, it is relevant to know the fraction of disorder while in others it is more important to know the exact position of the Disordered protein in the sequence. Since disorder can be used as a proxy either to estimate the complexity of an organism or complement a sequence search, it is also important for a predictor to be sufficiently disordered protein for genome-scale application.
For CAID, we report the maximum F 1-score F max —that is, maximum harmonic mean between precision and recall across all thresholdswhich takes into account predictions across the entire sensitivity spectrum ]
One thought on “Disordered protein”